spacepy.toolbox¶
Toolbox of various functions and generic utilities.
Authors: Steve Morley, Jon Niehof, Brian Larsen, Josef Koller, Dan Welling Institution: Los Alamos National Laboratory Contact: smorley@lanl.gov, jniehof@lanl.gov, balarsen@lanl.gov, jkoller@lanl.gov, dwelling@lanl.gov Los Alamos National Laboratory
Copyright 2010 Los Alamos National Security, LLC.
Functions
|
Split a sequence into subsequences based on value. |
|
assembles all pickled files matching fln_pattern into single file and save as outfln. |
|
Calculates bin width and number of bins for histogram using Freedman-Diaconis rule, if rule fails, defaults to square-root method |
|
Convert a list of bin centers to their edges |
|
Convert a list of bin edges to their centers |
|
Bootstrap confidence intervals for a histogram. |
|
pretty print a dictionary tree |
|
Convert a probability distribution function to a list of values |
|
Execute a function (or method) with a timeout. |
|
Times an event then prints out the time and the name of the event, nice for debugging and seeing that the code is progressing |
|
Returns geometrically spaced numbers. |
|
Return the full path of a parent directory with name as the leaf |
|
Read data from a URL |
|
Sort the given list in the way that humans expect. |
|
compute the N-dimensional hypot of an iterable or many arguments |
|
return the start and end indices implied by a range, useful when range is zero-length |
|
1-D linear interpolation with interpolation of hours/longitude |
|
given two array-like variables interweave them together. |
|
Find the function input such that definite integral is desired value. |
|
Returns if an object is a view of another object. |
|
Returns linear-spaced bins. |
|
load a pickle and return content as dictionary |
|
Returns log-spaced bins. |
|
Calculate median absolute deviation of a given input series |
|
Convert mlt values to radians for polar plotting transform mlt angles to radians from -pi to pi referenced from noon by default |
|
Given an input vector normalize the vector to a given range |
|
print min and max of input arrays |
|
Fit a Poisson distribution to data using the method and initial guess provided. |
|
print a progress bar with urllib.urlretrieve reporthook functionality |
|
Ask a yes/no question via raw_input() and return their answer. |
|
Convert radians values to mlt transform radians from -pi to pi to mlt referenced from noon by default |
|
save dictionary variable dict to a pickle with filename fln |
|
Finds the elements in a list of datetime objects present in another |
|
Finds the overlapping elements in two lists of datetime objects |
|
Find overlapping elements in two lists of datetime objects |
|
Split a job into subjobs and run a thread for each |
|
Apply a function to every element of a list, in separate threads |
|
Call a subprocess with a timeout. |
|
Given a multidimensional input return the unique rows or columns along the given axis. |
|
Download and update local database for omni, leapsecs etc |
|
Windowing mean function, window overlap is user defined |
Classes
|
Finds all links in a HTML page, useful for crawling. |
Exceptions
Raised when a time-limited process times out |
- spacepy.toolbox.arraybin(array, bins)[source]¶
Split a sequence into subsequences based on value.
Given a sequence of values and a sequence of values representing the division between bins, return the indices grouped by bin.
- Parameters:
- arrayarray_like
the input sequence to slice, must be sorted in ascending order
- binsarray_like
- dividing lines between bins. Number of bins is len(bins)+1,
value that exactly equal a dividing value are assigned to the higher bin
- Returns:
- outlist
indices for each bin (list of lists)
Examples
>>> import spacepy.toolbox as tb >>> tb.arraybin(range(10), [4.2]) [[0, 1, 2, 3, 4], [5, 6, 7, 8, 9]]
- spacepy.toolbox.assemble(fln_pattern, outfln, sortkey='ticks', verbose=True)[source]¶
assembles all pickled files matching fln_pattern into single file and save as outfln. Pattern may contain simple shell-style wildcards *? a la fnmatch file will be assembled along time axis given by Ticktock (key: ‘ticks’) in dictionary If sortkey = None, then nothing will be sorted
- Parameters:
- fln_patternstring
pattern to match filenames
- outflnstring
filename to save combined files to
- Returns:
- outdict
dictionary with combined values
Examples
>>> import spacepy.toolbox as tb >>> a, b, c = {'ticks':[1,2,3]}, {'ticks':[4,5,6]}, {'ticks':[7,8,9]} >>> tb.savepickle('input_files_2001.pkl', a) >>> tb.savepickle('input_files_2002.pkl', b) >>> tb.savepickle('input_files_2004.pkl', c) >>> a = tb.assemble('input_files_*.pkl', 'combined_input.pkl') ('adding ', 'input_files_2001.pkl') ('adding ', 'input_files_2002.pkl') ('adding ', 'input_files_2004.pkl') ('\n writing: ', 'combined_input.pkl') >>> print(a) {'ticks': array([1, 2, 3, 4, 5, 6, 7, 8, 9])}
- spacepy.toolbox.binHisto(data, verbose=False)[source]¶
Calculates bin width and number of bins for histogram using Freedman-Diaconis rule, if rule fails, defaults to square-root method
- The Freedman-Diaconis method is detailed in:
Freedman, D., and P. Diaconis (1981), On the histogram as a density estimator: L2 theory, Z. Wahrscheinlichkeitstheor. Verw. Geb., 57, 453–476
- and is also described by:
Wilks, D. S. (2006), Statistical Methods in the Atmospheric Sciences, 2nd ed.
- Parameters:
- dataarray_like
list/array of data values
- verboseboolean (optional)
print out some more information
- Returns:
- outtuple
calculated width of bins using F-D rule, number of bins (nearest integer) to use for histogram
See also
Examples
>>> import numpy, spacepy >>> import matplotlib.pyplot as plt >>> numpy.random.seed(8675301) >>> data = numpy.random.randn(1000) >>> binw, nbins = spacepy.toolbox.binHisto(data) >>> print(nbins) 19 >>> p = plt.hist(data, bins=nbins, histtype='step', density=True)
- spacepy.toolbox.bin_center_to_edges(centers)[source]¶
Convert a list of bin centers to their edges
Given a list of center values for a set of bins, finds the start and end value for each bin. (start of bin n+1 is assumed to be end of bin n). Useful for e.g. matplotlib.pyplot.pcolor.
Edge between bins n and n+1 is arithmetic mean of the center of n and n+1; edge below bin 0 and above last bin are established to make these bins symmetric about their center value.
- Parameters:
- centerslist
list of center values for bins
- Returns:
- outlist
list of edges for bins
- note: returned list will be one element longer than centers
Examples
>>> import spacepy.toolbox as tb >>> tb.bin_center_to_edges([1,2,3]) [0.5, 1.5, 2.5, 3.5]
- spacepy.toolbox.bin_edges_to_center(edges)[source]¶
Convert a list of bin edges to their centers
Given a list of edge values for a set of bins, finds the center of each bin. (start of bin n+1 is assumed to be end of bin n).
Center of bin n is arithmetic mean of the edges of the adjacent bins.
- Parameters:
- edgeslist
list of edge values for bins
- Returns:
- outnumpy.ndarray
array of centers for bins
- note: returned array will be one element shorter than edges
Examples
>>> import spacepy.toolbox as tb >>> tb.bin_center_to_edges([1,2,3]) [0.5, 1.5, 2.5, 3.5]
- spacepy.toolbox.bootHisto(data, inter=90.0, n=1000, seed=None, plot=False, target=None, figsize=None, loc=None, **kwargs)[source]¶
Bootstrap confidence intervals for a histogram.
All other keyword arguments are passed to
numpy.histogram()ormatplotlib.pyplot.bar().Changed in version 0.2.3: This argument pass-through did not work in earlier versions of SpacePy.
- Parameters:
- dataarray_like
list/array of data values
- interfloat (optional; default 90)
percentage confidence interval to return. Default 90% (i.e. lower CI will be 5% and upper will be 95%)
- nint (optional; default 1000)
number of bootstrap iterations
- seedint (optional)
Optional seed for the random number generator. If not specified; numpy generator will not be reseeded.
- plotbool (optional)
Plot the result. Plots if True or
target,figsize, orlocspecified.- target(optional)
Target on which to plot the figure (figure or axes). See
spacepy.plot.utils.set_target()for details.- figsizetuple (optional)
Passed to
spacepy.plot.utils.set_target().- locint (optional)
Passed to
spacepy.plot.utils.set_target().
- Returns:
- outtuple
tuple of bin_edges, low, high, sample[, bars]. Where
bin_edgesis the edges of the bins used;lowis the histogram with the value for each bin from the bottom of that bin’s confidence interval;highsimilarly for the top;sampleis the histogram of the input sample without resampling. If plotting, also returned isbars, the container object returned from matplotlib.
Notes
Added in version 0.2.1.
The confidence intervals are calculated for each bin individually and thus the resulting low/high histograms may not have actually occurred in the calculation from the surrogates. If using a probability density histogram, this can have “interesting” implications for interpretation.
Examples
>>> import numpy.random >>> import spacepy.toolbox >>> numpy.random.seed(0) >>> data = numpy.random.randn(1000) >>> bin_edges, low, high, sample, bars = spacepy.toolbox.bootHisto( ... data, plot=True)
(
Source code,png,hires.png,pdf)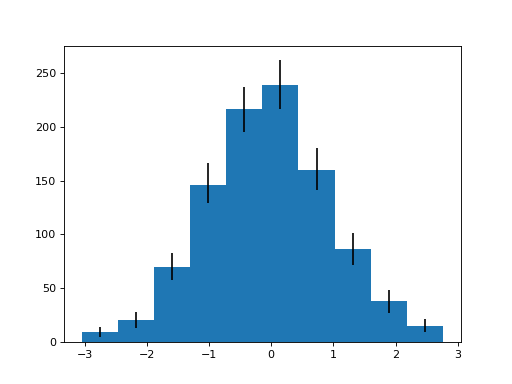
- spacepy.toolbox.dictree(in_dict, verbose=False, spaces=None, levels=True, attrs=False, print_out=True, **kwargs)[source]¶
pretty print a dictionary tree
- Parameters:
- in_dictdict
a complex dictionary (with substructures)
- verbosebool, default False
print more info
- spacesstr (optional)
string will added for every line
- levelsint (optional)
number of levels to recurse through (True, the default, means all)
- attrsbool, default False
display information for attributes
- print_outbool, default True
Added in version 0.5.0.
Print output (original behavior); if
False, return the output.
- Raises:
- TypeError
Input does not have keys or attrs, cannot build tree.
Examples
>>> import spacepy.toolbox as tb >>> d = {'grade':{'level1':[4,5,6], 'level2':[2,3,4]}, 'name':['Mary', 'John', 'Chris']} >>> tb.dictree(d) + |____grade |____level1 |____level2 |____name
More complicated example using a datamodel:
>>> from spacepy import datamodel >>> counts = datamodel.dmarray([2,4,6], attrs={'units': 'cts/s'}) >>> data = {'counts': counts, 'PI': 'Dr Zog'} >>> tb.dictree(data) + |____PI |____counts >>> tb.dictree(data, attrs=True, verbose=True) + |____PI (str [6]) |____counts (spacepy.datamodel.dmarray (3,)) :|____units (str [5])
Attributes of, e.g., a CDF or a datamodel type object (obj.attrs) are denoted by a colon.
- spacepy.toolbox.dist_to_list(func, length, min=None, max=None)[source]¶
Convert a probability distribution function to a list of values
This is a deterministic way to produce a known-length list of values matching a certain probability distribution. It is likely to be a closer match to the distribution function than a random sampling from the distribution.
- Parameters:
- funccallable
- function to call for each possible value, returning
probability density at that value (does not need to be normalized.)
- lengthint
number of elements to return
- minfloat
minimum value to possibly include
- maxfloat
maximum value to possibly include
Examples
>>> import matplotlib >>> import numpy >>> import spacepy.toolbox as tb >>> gauss = lambda x: math.exp(-(x ** 2) / (2 * 5 ** 2)) / (5 * math.sqrt(2 * math.pi)) >>> vals = tb.dist_to_list(gauss, 1000, -numpy.inf, numpy.inf) >>> print vals[0] -16.45263... >>> p1 = matplotlib.pyplot.hist(vals, bins=[i - 10 for i in range(21)], facecolor='green') >>> matplotlib.pyplot.hold(True) >>> x = [i / 100.0 - 10.0 for i in range(2001)] >>> p2 = matplotlib.pyplot.plot(x, [gauss(i) * 1000 for i in x], 'red') >>> matplotlib.pyplot.draw()
- spacepy.toolbox.do_with_timeout(timeout, target, *args, **kwargs)[source]¶
Execute a function (or method) with a timeout.
Call the function (or method)
target, with argumentsargsand keyword argumentskwargs. Normally return the return value fromtarget, but iftargettakes more thantimeoutseconds to execute, raisesTimeoutError.Note
This is, at best, a blunt instrument. Exceptions from
targetmay not propagate properly (tracebacks will be hard to follow.) The function which failed to time out may continue to execute until the interpreter exits; trapping the TimeoutError and continuing normally is not recommended.- Parameters:
- timeoutfloat
Timeout, in seconds.
- targetcallable
- Python callable (generally a function, may also be an
imported ctypes function) to run.
- argssequence
Arguments to pass to
target.- kwargsdict
keyword arguments to pass to
target.
- Returns:
- out
return value of
target
- Raises:
- TimeoutError
If
targetdoes not return intimeoutseconds.
Examples
>>> import spacepy.toolbox as tb >>> import time >>> def time_me_out(): ... time.sleep(5) >>> tb.do_with_timeout(0.5, time_me_out) #raises TimeoutError
- spacepy.toolbox.eventTimer(Event, Time1)[source]¶
Times an event then prints out the time and the name of the event, nice for debugging and seeing that the code is progressing
- Parameters:
- Eventstr
Name of the event, string is printed out by function
- Time1time.time
the time to difference in the function
- Returns:
- Time2time.time
the new time for the next call to EventTimer
Examples
>>> import spacepy.toolbox as tb >>> import time >>> t1 = time.time() >>> t1 = tb.eventTimer('Test event finished', t1) ('4.40', 'Test event finished')
- spacepy.toolbox.geomspace(start, ratio=None, stop=False, num=50)[source]¶
Returns geometrically spaced numbers.
- Parameters:
- startfloat
The starting value of the sequence.
- ratiofloat (optional)
The ratio between subsequent points
- stopfloat (optional)
End value, if this is selected
numis overridden- numint (optional)
Number of samples to generate. Default is 50.
- Returns:
- seqarray
geometrically spaced sequence
Examples
To get a geometric progression between 0.01 and 3 in 10 steps
>>> import spacepy.toolbox as tb >>> tb.geomspace(0.01, stop=3, num=10) [0.01, 0.018846716378431192, 0.035519871824902655, 0.066943295008216955, 0.12616612944575134, 0.23778172582285118, 0.44814047465571644, 0.84459764235318191, 1.5917892219322083, 2.9999999999999996]
To get a geometric progression with a specified ratio, say 10
>>> import spacepy.toolbox as tb >>> tb.geomspace(0.01, ratio=10, num=5) [0.01, 0.10000000000000001, 1.0, 10.0, 100.0]
- spacepy.toolbox.getNamedPath(name)[source]¶
Return the full path of a parent directory with name as the leaf
- Parameters:
- namestring
the name of the parent directory to locate
Examples
Run from a directory /mnt/projects/dream/bin/Ephem with ‘dream’ as the name, this function would return ‘/mnt/projects/dream’
- spacepy.toolbox.get_url(url, outfile=None, reporthook=None, cached=False, keepalive=False, conn=None)[source]¶
Read data from a URL
Open an HTTP URL, honoring the user agent as specified in the SpacePy config file. Returns the data, optionally also writing out to a file.
This is similar to the deprecated
urlretrieve.Changed in version 0.5.0: In earlier versions of SpacePy invalid combinations of cached and outfile raised RuntimeError, changed to ValueError.
- Parameters:
- urlstr
The URL to open
- outfilestr (optional)
Full path to file to write data to
- reporthookcallable (optional)
Function for reporting progress; takes arguments of block count, block size, and total size.
- cachedbool (optional)
Compare modification time of the URL to the modification time of
outfile; do not retrieve (and return None) unless the URL is newer than the file. If set outfile is required.- keepalivebool (optional)
Attempt to keep the connection open to retrieve more URLs. The return becomes a tuple of (data, conn) to return the connection used so it can be used again. This mode does not support proxies. Required to be True if conn is provided. (Default False)
- connhttp.client.HTTPConnection (optional)
An established http connection (HTTPS is also okay) to use with
keepalive. If not provided, will attempt to make a connection.
- Returns:
- bytes
The HTTP data from the server.
See also
Notes
This function honors proxy settings as described in
urllib.request.getproxies(). Cryptic error messages (such asNetwork is unreachable) may indicate that proxy settings should be defined as appropriate for your environment (e.g. withHTTP_PROXYorHTTPS_PROXYenvironment variables).
- spacepy.toolbox.human_sort(l)[source]¶
Sort the given list in the way that humans expect. http://www.codinghorror.com/blog/2007/12/sorting-for-humans-natural-sort-order.html
- Parameters:
- llist
list of objects to human sort
- Returns:
- outlist
sorted list
Examples
>>> import spacepy.toolbox as tb >>> dat = ['r1.txt', 'r10.txt', 'r2.txt'] >>> dat.sort() >>> print dat ['r1.txt', 'r10.txt', 'r2.txt'] >>> tb.human_sort(dat) ['r1.txt', 'r2.txt', 'r10.txt']
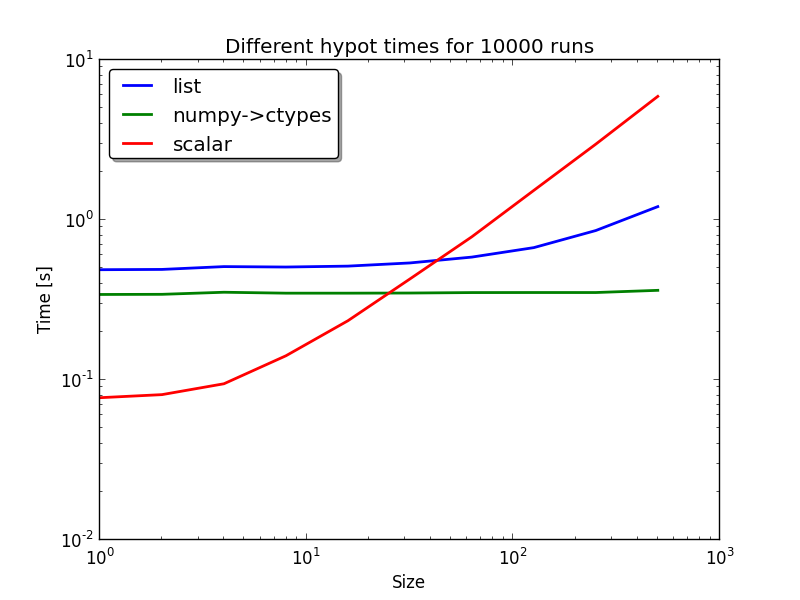
- spacepy.toolbox.hypot(*args)[source]¶
compute the N-dimensional hypot of an iterable or many arguments
- Parameters:
- argsmany numbers or array-like
array like or many inputs to compute from
- Returns:
- outfloat
N-dimensional hypot of a number
Notes
- This function has a complicated speed function.
if a numpy array of floats is input this is passed off to C
if iterables are passed in they are made into numpy arrays and comptaton is done local
if many scalar agruments are passed in calculation is done in a loop
- For max speed:
- <20 elements expand them into scalars
>>> tb.hypot(*vals) >>> tb.hypot(vals[0], vals[1]...) #alternate
>20 elements premake them into a numpy array of doubles
Examples
>>> from spacepy import toolbox as tb >>> print tb.hypot([3,4]) 5.0 >>> print tb.hypot(3,4) 5.0 >>> # Benchmark #### >>> from spacepy import toolbox as tb >>> import numpy as np >>> import timeit >>> num_list = [] >>> num_np = [] >>> num_np_double = [] >>> num_scalar = [] >>> tot = 500 >>> for num in tb.logspace(1, tot, 10): >>> print num >>> num_list.append(timeit.timeit(stmt='tb.hypot(a)', setup='from spacepy import toolbox as tb; import numpy as np; a = [3]*{0}'.format(int(num)), number=10000)) >>> num_np.append(timeit.timeit(stmt='tb.hypot(a)', setup='from spacepy import toolbox as tb; import numpy as np; a = np.asarray([3]*{0})'.format(int(num)), number=10000)) >>> num_scalar.append(timeit.timeit(stmt='tb.hypot(*a)', setup='from spacepy import toolbox as tb; import numpy as np; a = [3]*{0}'.format(int(num)), number=10000)) >>> from pylab import * >>> loglog(tb.logspace(1, tot, 10), num_list, lw=2, label='list') >>> loglog(tb.logspace(1, tot, 10), num_np, lw=2, label='numpy->ctypes') >>> loglog(tb.logspace(1, tot, 10), num_scalar, lw=2, label='scalar') >>> legend(shadow=True, fancybox=1, loc='upper left') >>> title('Different hypot times for 10000 runs') >>> ylabel('Time [s]') >>> xlabel('Size')
- spacepy.toolbox.indsFromXrange(inxrange)[source]¶
return the start and end indices implied by a range, useful when range is zero-length
- Parameters:
- inxrangerange
input range object to parse
- Returns:
- list of int
List of start, stop indices in the range. The return value is not defined if a stride is specified or if stop is before start (but will work when stop equals start).
Examples
>>> import spacepy.toolbox as tb >>> foo = range(23, 39) >>> foo[0] 23 >>> tb.indsFromXrange(foo) [23, 39] >>> foo1 = range(23, 23) >>> tb.indsFromXrange(foo) #indexing won't work in this case [23, 23]
- spacepy.toolbox.interpol(newx, x, y, wrap=None, **kwargs)[source]¶
1-D linear interpolation with interpolation of hours/longitude
- Parameters:
- newxarray_like
x values where we want the interpolated values
- xarray_like
x values of the original data (must be monotonically increasing or wrapping)
- yarray_like
y values of the original data
- wrapstring, optional
for continuous x data that wraps in y at ‘hours’ (24), ‘longitude’ (360), or arbitrary value (int, float)
- kwargsdict
additional keywords, currently accepts baddata that sets baddata for masked arrays
- Returns:
- outnumpy.masked_array
interpolated data values for new abscissa values
Examples
For a simple interpolation
>>> import spacepy.toolbox as tb >>> import numpy >>> x = numpy.arange(10) >>> y = numpy.arange(10) >>> tb.interpol(numpy.arange(5)+0.5, x, y) array([ 0.5, 1.5, 2.5, 3.5, 4.5])
To use the wrap functionality, without the wrap keyword you get the wrong answer
>>> y = range(24)*2 >>> x = range(len(y)) >>> tb.interpol([1.5, 10.5, 23.5], x, y, wrap='hour').compressed() # compress removed the masked array array([ 1.5, 10.5, 23.5]) >>> tb.interpol([1.5, 10.5, 23.5], x, y) array([ 1.5, 10.5, 11.5])
- spacepy.toolbox.interweave(a, b)[source]¶
given two array-like variables interweave them together. Discussed here: http://stackoverflow.com/questions/5347065/interweaving-two-numpy-arrays
- Parameters:
- aarray-like
first array
- barray-like
second array
- Returns:
- outnumpy.ndarray
interweaved array
- spacepy.toolbox.intsolve(func, value, start=None, stop=None, maxit=1000)[source]¶
Find the function input such that definite integral is desired value.
Given a function, integrate from an (optional) start point until the integral reached a desired value, and return the end point of the integration.
- Parameters:
- funccallable
function to integrate, must take single parameter
- valuefloat
desired final value of the integral
- startfloat (optional)
value at which to start integration, default -Infinity
- stopfloat (optional)
value at which to stop integration, default +Infinity
- maxitinteger
maximum number of iterations
- Returns:
- outfloat
x such that the integral of L{func} from L{start} to x is L{value}
- Note: Assumes func is everywhere positive, otherwise solution may
be multi-valued.
- spacepy.toolbox.isview(array1, array2=None)[source]¶
Returns if an object is a view of another object. More precisely if one array argument is specified True is returned is the arrays owns its data. If two arrays arguments are specified a tuple is returned of if the first array owns its data and the the second if they point at the same memory location
- Parameters:
- array1numpy.ndarray
array to query if it owns its data
- Returns:
- outbool or tuple
If one array is specified bool is returned, True is the array owns its data. If two arrays are specified a tuple where the second element is a bool of if the array point at the same memory location
- Other Parameters:
- array2object (optional)
array to query if array1 is a view of this object at the specified memory location
Examples
import numpy import spacepy.toolbox as tb a = numpy.arange(100) b = a[0:10] tb.isview(a) # False tb.isview(b) # True tb.isview(b, a) # (True, True) tb.isview(b, b) # (True, True) # the conditions are met and numpy cannot tell this
- spacepy.toolbox.linspace(min, max, num, **kwargs)[source]¶
Returns linear-spaced bins. Same as numpy.linspace except works with datetime and is faster
- Parameters:
- minfloat, datetime
minimum value
- maxfloat, datetime
maximum value
- numinteger
number of linear spaced bins
- Returns:
- outarray
linear-spaced bins from min to max in a numpy array
- Other Parameters:
- kwargsdict
additional keywords passed into matplotlib.dates.num2date
Notes
This function works on both numbers and datetime objects. Not leapsecond aware.
Examples
>>> import spacepy.toolbox as tb >>> tb.linspace(1, 10, 4) array([ 1., 4., 7., 10.])
- spacepy.toolbox.loadpickle(fln)[source]¶
load a pickle and return content as dictionary
- Parameters:
- flnstring
filename
- Returns:
- outdict
dictionary with content from file
See also
Examples
- note: If fln is not found, but the same filename with ‘.gz’
is found, will attempt to open the .gz as a gzipped file.
>>> d = loadpickle('test.pbin')
- spacepy.toolbox.logspace(min, max, num, **kwargs)[source]¶
Returns log-spaced bins. Same as numpy.logspace except the min and max are the min and max not log10(min) and log10(max)
- Parameters:
- minfloat
minimum value
- maxfloat
maximum value
- numinteger
number of log spaced bins
- Returns:
- outarray
log-spaced bins from min to max in a numpy array
- Other Parameters:
- kwargsdict
additional keywords passed into matplotlib.dates.num2date
Notes
This function works on both numbers and datetime objects. Not leapsecond aware.
Examples
>>> import spacepy.toolbox as tb >>> tb.logspace(1, 100, 5) array([ 1. , 3.16227766, 10. , 31.6227766 , 100. ])
- spacepy.toolbox.medAbsDev(series, scale=False)[source]¶
Calculate median absolute deviation of a given input series
Median absolute deviation (MAD) is a robust and resistant measure of the spread of a sample (same purpose as standard deviation). The MAD is preferred to the inter-quartile range as the inter-quartile range only shows 50% of the data whereas the MAD uses all data but remains robust and resistant. See e.g. Wilks, Statistical methods for the Atmospheric Sciences, 1995, Ch. 3. For additional details on the scaling, see Rousseeuw and Croux, J. Amer. Stat. Assoc., 88 (424), pp. 1273-1283, 1993.
- Parameters:
- seriesarray_like
the input data series
- Returns:
- outfloat
the median absolute deviation
- Other Parameters:
- scalebool
if True (default: False), scale to standard deviation of a normal distribution
Examples
Find the median absolute deviation of a data set. Here we use the log- normal distribution fitted to the population of sawtooth intervals, see Morley and Henderson, Comment, Geophysical Research Letters, 2009.
>>> import numpy >>> import spacepy.toolbox as tb >>> numpy.random.seed(8675301) >>> data = numpy.random.lognormal(mean=5.1458, sigma=0.302313, size=30) >>> print data array([ 181.28078923, 131.18152745, ... , 141.15455416, 160.88972791]) >>> tb.medAbsDev(data) 28.346646721370192
note This implementation is robust to presence of NaNs
- spacepy.toolbox.mlt2rad(mlt, midnight=False)[source]¶
Convert mlt values to radians for polar plotting transform mlt angles to radians from -pi to pi referenced from noon by default
- Parameters:
- mltnumpy array
array of mlt values
- midnightboolean (optional)
reference to midnight instead of noon
- Returns:
- outnumpy array
array of radians
See also
Examples
>>> from numpy import array >>> mlt2rad(array([3,6,9,14,22])) array([-2.35619449, -1.57079633, -0.78539816, 0.52359878, 2.61799388])
- spacepy.toolbox.normalize(vec, low=0.0, high=1.0)[source]¶
Given an input vector normalize the vector to a given range
- Parameters:
- vecarray_like
input vector to normalize
- lowfloat
minimum value to scale to, default 0.0
- highfloat
maximum value to scale to, default 1.0
- Returns:
- outarray_like
normalized vector
Examples
>>> import spacepy.toolbox as tb >>> tb.normalize([1,2,3]) [0.0, 0.5, 1.0]
- spacepy.toolbox.pmm(*args)[source]¶
print min and max of input arrays
- Parameters:
- aarray-like
arbitrary number of input arrays (or lists)
- Returns:
- outlist
list of min, max for each array
Examples
>>> import spacepy.toolbox as tb >>> from numpy import arange >>> tb.pmm(arange(10), arange(10)+3) [[0, 9], [3, 12]]
- spacepy.toolbox.poisson_fit(data, initial=None, method='Powell')[source]¶
Fit a Poisson distribution to data using the method and initial guess provided.
- Parameters:
- dataarray-like
Data to fit a Poisson distribution to.
- initialint or None
initial guess for the fit, if None np.median(data) is used
- methodstr
method passed to scipy.optimize.minimize, default=’Powell’
- Returns:
- resultscipy.optimize.optimize.OptimizeResult
Resulting fit results from scipy.optimize, answer is result.x, user should likely round.
Examples
>>> import spacepy.toolbox as tb >>> from scipy.stats import poisson >>> import matplotlib.pyplot as plt >>> import numpy as np >>> data = poisson.rvs(20, size=1000) >>> res = tb.poisson_fit(data) >>> print(res.x) 19.718000038769095 >>> xvals = np.arange(0, np.max(data)+5) >>> plt.hist(data, bins=xvals, normed=True) >>> plt.plot(xvals, poisson.pmf(xvals, np.round(res.x)))
- spacepy.toolbox.progressbar(count, blocksize, totalsize, text='Download Progress')[source]¶
print a progress bar with urllib.urlretrieve reporthook functionality
Examples
>>> import spacepy.toolbox as tb >>> import urllib >>> urllib.urlretrieve(config['psddata_url'], PSDdata_fname, reporthook=tb.progressbar)
- spacepy.toolbox.query_yes_no(question, default='yes')[source]¶
Ask a yes/no question via raw_input() and return their answer.
“question” is a string that is presented to the user. “default” is the presumed answer if the user just hits <Enter>. It must be “yes” (the default), “no” or None (meaning an answer is required of the user).
The “answer” return value is one of “yes” or “no”.
- Parameters:
- questionstr
the question to ask
- defaultstr (optional)
- Returns:
- outstr
answer (‘yes’ or ‘no’)
- Raises:
- ValueError
The default answer is not in (None|”yes”|”no”)
Examples
>>> import spacepy.toolbox as tb >>> tb.query_yes_no('Ready to go?') Ready to go? [Y/n] y 'yes'
- spacepy.toolbox.rad2mlt(rad, midnight=False)[source]¶
Convert radians values to mlt transform radians from -pi to pi to mlt referenced from noon by default
- Parameters:
- radnumpy array
array of radian values
- midnightboolean (optional)
reference to midnight instead of noon
- Returns:
- outnumpy array
array of mlt values
See also
Examples
>>> rad2mlt(array([0,pi, pi/2.])) array([ 12., 24., 18.])
- spacepy.toolbox.savepickle(fln, dict, compress=None)[source]¶
save dictionary variable dict to a pickle with filename fln
- Parameters:
- flnstring
filename
- dictdict
container with stuff
- compressbool
- write as a gzip-compressed file
(.gz will be added to
fln). If not specified, defaults to uncompressed, unless the compressed file exists and the uncompressed does not.
See also
Examples
>>> d = {'grade':[1,2,3], 'name':['Mary', 'John', 'Chris']} >>> savepickle('test.pbin', d)
- spacepy.toolbox.tCommon(ts1, ts2, mask_only=True)[source]¶
Finds the elements in a list of datetime objects present in another
- Parameters:
- ts1list or array-like
first set of datetime objects
- ts2list or array-like
second set of datetime objects
- Returns:
- outtuple
Two element tuple of truth tables (of 1 present in 2, & vice versa)
See also
Examples
>>> import spacepy.toolbox as tb >>> import numpy as np >>> import datetime as dt >>> ts1 = np.array([dt.datetime(2001,3,10)+dt.timedelta(hours=a) for a in range(20)]) >>> ts2 = np.array([dt.datetime(2001,3,10,2)+dt.timedelta(hours=a*0.5) for a in range(20)]) >>> common_inds = tb.tCommon(ts1, ts2) >>> common_inds[0] #mask of values in ts1 common with ts2 array([False, False, True, True, True, True, True, True, True, True, True, True, False, False, False, False, False, False, False, False], dtype=bool) >>> ts2[common_inds[1]] #values of ts2 also in ts1
The latter can be found more simply by setting the mask_only keyword to False
>>> common_vals = tb.tCommon(ts1, ts2, mask_only=False) >>> common_vals[1] array([2001-03-10 02:00:00, 2001-03-10 03:00:00, 2001-03-10 04:00:00, 2001-03-10 05:00:00, 2001-03-10 06:00:00, 2001-03-10 07:00:00, 2001-03-10 08:00:00, 2001-03-10 09:00:00, 2001-03-10 10:00:00, 2001-03-10 11:00:00], dtype=object)
- spacepy.toolbox.tOverlap(ts1, ts2, *args, **kwargs)[source]¶
Finds the overlapping elements in two lists of datetime objects
- Parameters:
- ts1datetime
first set of datetime object
- ts2datetime
datatime object
- args
additional arguments passed to tOverlapHalf
- Returns:
- outlist
indices of ts1 within interval of ts2, & vice versa
See also
Examples
Given two series of datetime objects, event_dates and omni[‘Time’]:
>>> import spacepy.toolbox as tb >>> from spacepy import omni >>> import datetime >>> event_dates = st.tickrange(datetime.datetime(2000, 1, 1), datetime.datetime(2000, 10, 1), deltadays=3) >>> onni_dates = st.tickrange(datetime.datetime(2000, 1, 1), datetime.datetime(2000, 10, 1), deltadays=0.5) >>> omni = omni.get_omni(onni_dates) >>> [einds,oinds] = tb.tOverlap(event_dates, omni['ticks']) >>> omni_time = omni['ticks'][oinds[0]:oinds[-1]+1] >>> print omni_time [datetime.datetime(2000, 1, 1, 0, 0), datetime.datetime(2000, 1, 1, 12, 0), ... , datetime.datetime(2000, 9, 30, 0, 0)]
- spacepy.toolbox.tOverlapHalf(ts1, ts2, presort=False)[source]¶
Find overlapping elements in two lists of datetime objects
This is one-half of tOverlap, i.e. it finds only occurrences where ts2 exists within the bounds of ts1, or the second element returned by tOverlap.
- Parameters:
- ts1list
first set of datetime object
- ts2list
datatime object
- presortbool
- Set to use a faster algorithm which assumes ts1 and
ts2 are both sorted in ascending order. This speeds up the overlap comparison by about 50x, so it is worth sorting the list if one sort can be done for many calls to tOverlap
- Returns:
- outlist
indices of ts2 within interval of ts1
note: Returns empty list if no overlap found
- spacepy.toolbox.thread_job(job_size, thread_count, target, *args, **kwargs)[source]¶
Split a job into subjobs and run a thread for each
Each thread spawned will call L{target} to handle a slice of the job.
- This is only useful if a job:
Can be split into completely independent subjobs
Relies heavily on code that does not use the Python GIL, e.g. numpy or ctypes code
Does not return a value. Either pass in a list/array to hold the result, or see L{thread_map}
- Parameters:
- job_sizeint
Total size of the job. Often this is an array size.
- thread_countint
- Number of threads to spawn. If =0 or None, will
spawn as many threads as there are cores available on the system. (Each hyperthreading core counts as 2.) Generally this is the Right Thing to do. If NEGATIVE, will spawn abs(thread_count) threads, but will run them sequentially rather than in parallel; useful for debugging.
- targetcallable
- Python callable (generally a function, may also be an
imported ctypes function) to run in each thread. The last two positional arguments passed in will be a “start” and a “subjob size,” respectively; frequently this will be the start index and the number of elements to process in an array.
- argssequence
- Arguments to pass to L{target}. If L{target} is an instance
method, self must be explicitly passed in. start and subjob_size will be appended.
- kwargsdict
keyword arguments to pass to L{target}.
Examples
squaring 100 million numbers:
>>> import numpy >>> import spacepy.toolbox as tb >>> numpy.random.seed(8675301) >>> a = numpy.random.randint(0, 100, [100000000]) >>> b = numpy.empty([100000000], dtype='int64') >>> def targ(in_array, out_array, start, count): out_array[start:start + count] = in_array[start:start + count] ** 2 >>> tb.thread_job(len(a), 0, targ, a, b) >>> print(b[0:5]) [2704 7225 196 1521 36]
- This example:
Defines a target function, which will be called for each thread. It is usually necessary to define a simple “wrapper” function like this to provide the correct call signature.
The target function receives inputs C{in_array} and C{out_array}, which are not touched directly by C{thread_job} but are passed through in the call. In this case, C{a} gets passed as C{in_array} and C{b} as C{out_array}
The target function also receives the start and number of elements it needs to process. For each thread where the target is called, these numbers are different.
- spacepy.toolbox.thread_map(target, iterable, thread_count=None, *args, **kwargs)[source]¶
Apply a function to every element of a list, in separate threads
Interface is similar to multiprocessing.map, except it runs in threads
This is made largely obsolete in python3 by from concurrent import futures
- Parameters:
- targetcallable
- Python callable to run on each element of iterable.
For each call, an element of iterable is appended to args and both args and kwargs are passed through. Note that this means the iterable element is always the last positional argument; this allows the specification of self as the first argument for method calls.
- iterableiterable
elements to pass to each call of L{target}
- argssequence
- arguments to pass to target before each element of
iterable
- thread_countinteger
Number of threads to spawn; see L{thread_job}.
- kwargsdict
keyword arguments to pass to L{target}.
- Returns:
- outlist
return values of L{target} for each item from L{iterable}
Examples
find totals of several arrays
>>> import numpy >>> from spacepy import toolbox >>> inputs = range(100) >>> totals = toolbox.thread_map(numpy.sum, inputs) >>> print(totals[0], totals[50], totals[99]) (0, 50, 99)
>>> # in python3 >>> from concurrent import futures >>> with futures.ThreadPoolExecutor(max_workers=4) as executor: ...: for ans in executor.map(numpy.sum, [0,50,99]): ...: print ans #0 #50 #99
- spacepy.toolbox.timeout_check_call(timeout, *args, **kwargs)[source]¶
Call a subprocess with a timeout.
Deprecated since version 0.7.0: Use
timeoutargument ofsubprocess.check_call(), added in Python 3.3.Like
subprocess.check_call(), but will terminate the process and raiseTimeoutErrorif it runs for too long.This will only terminate the single process started; any child processes will remain running (this has implications for, say, spawing shells.)
- Parameters:
- timeoutfloat
Timeout, in seconds. Fractions are acceptable but the resolution is of order 100ms.
- argssequence
Arguments passed through to
subprocess.Popen- kwargsdict
keyword arguments to pass to
subprocess.Popen
- Returns:
- outint
0 on successful completion
- Raises:
- TimeoutError
If subprocess does not return in
timeoutseconds.- CalledProcessError
If command has non-zero exit status
Examples
>>> import spacepy.toolbox as tb >>> tb.timeout_check_call(1, 'sleep 30', shell=True) #raises TimeoutError
- spacepy.toolbox.unique_columns(inval, axis=0)[source]¶
Given a multidimensional input return the unique rows or columns along the given axis. Based largely on http://stackoverflow.com/questions/16970982/find-unique-rows-in-numpy-array axis=0 is unique rows, axis=1 is unique columns
- Parameters:
- invalarray-like
array to find unique columns or rows of
- Returns:
- outarray
N-dimensional array of the unique values along the axis
- Other Parameters:
- axisint
The axis to find unique over, default: 0
- spacepy.toolbox.update(all=True, QDomni=False, omni=False, omni2=False, leapsecs=False, PSDdata=False, cached=True)[source]¶
Download and update local database for omni, leapsecs etc
Web access is via
get_url(); notes there may be helpful in debugging errors. See also thekeepaliveconfiguration option.- Parameters:
- allboolean (optional)
if True, update OMNI2, Qin-Denton and leapsecs
- omniboolean (optional)
if True. update only omni (Qin-Denton)
- omni2boolean (optional)
if True, update only original OMNI2
- QDomniboolean (optional)
if True, update OMNI2 and Qin-Denton
- leapsecsboolean (optional)
if True, update only leapseconds
- cachedboolean (optional)
Only update files if timestamp on server is newer than timestamp on local file (default). Set False to always download files.
- Returns:
- outstring
data directory where things are saved
See also
Examples
>>> import spacepy.toolbox as tb >>> tb.update(omni=True)
- spacepy.toolbox.windowMean(data, time=[], winsize=0, overlap=0, st_time=None, op=<function mean>)[source]¶
Windowing mean function, window overlap is user defined
- Parameters:
- dataarray_like
1D series of points
- timelist (optional)
series of timestamps, optional (format as numeric or datetime) For non-overlapping windows set overlap to zero. Must be same length as data.
- winsizeinteger or datetime.timedelta (optional)
window size
- overlapinteger or datetime.timedelta (optional)
amount of window overlap
- st_timedatetime.datetime (optional)
for time-based averaging, a start-time other than the first point can be specified
- opcallable (optional)
the operator to be called, default numpy.mean
- Returns:
- outtuple
the windowed mean of the data, and an associated reference time vector
Examples
For non-overlapping windows set overlap to zero. e.g. (time-based averaging) Given a data set of 100 points at hourly resolution (with the time tick in the middle of the sample), the daily average of this, with half-overlapping windows is calculated:
>>> import spacepy.toolbox as tb >>> from datetime import datetime, timedelta >>> wsize = datetime.timedelta(days=1) >>> olap = datetime.timedelta(hours=12) >>> data = [10, 20]*50 >>> time = [datetime.datetime(2001,1,1) + datetime.timedelta(hours=n, minutes = 30) for n in range(100)] >>> outdata, outtime = tb.windowMean(data, time, winsize=wsize, overlap=olap, st_time=datetime.datetime(2001,1,1)) >>> outdata, outtime ([15.0, 15.0, 15.0, 15.0, 15.0, 15.0, 15.0], [datetime.datetime(2001, 1, 1, 12, 0), datetime.datetime(2001, 1, 2, 0, 0), datetime.datetime(2001, 1, 2, 12, 0), datetime.datetime(2001, 1, 3, 0, 0), datetime.datetime(2001, 1, 3, 12, 0), datetime.datetime(2001, 1, 4, 0, 0), datetime.datetime(2001, 1, 4, 12, 0)])
When using time-based averaging, ensure that the time tick corresponds to the middle of the time-bin to which the data apply. That is, if the data are hourly, say for 00:00-01:00, then the time applied should be 00:30. If this is not done, unexpected behaviour can result.
e.g. (pointwise averaging),
>>> outdata, outtime = tb.windowMean(data, winsize=24, overlap=12) >>> outdata, outtime ([15.0, 15.0, 15.0, 15.0, 15.0, 15.0, 15.0], [12.0, 24.0, 36.0, 48.0, 60.0, 72.0, 84.0])
where winsize and overlap are numeric, in this example the window size is 24 points (as the data are hourly) and the overlap is 12 points (a half day). The output vectors start at winsize/2 and end at N-(winsize/2), the output time vector is basically a reference to the nth point in the original series.
note This is a quick and dirty function - it is NOT optimized, at all.

